- Prepare samples and controls
- Reconstitute Magic Red by adding 50 µL DMSO.
- Dilute Magic Red 1:10 by adding 450 µL diH2O.
- Add 20 µL Magic Red to each sample (~ 500 µL aliquot of cultured cells).
- Incubate while protected from light.
- Watch color start to develop within 15 minutes.
- If desired, label with additional stains, such as Hoechst, DAPI, Acridine orange, or an antibody.
- If desired, fix or embed cells.
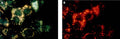
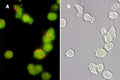
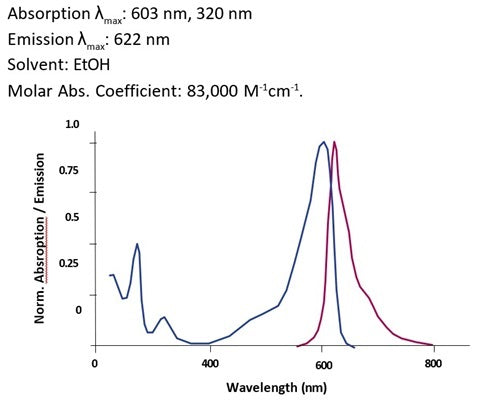
- Analyze with a fluorescence microscope or fluorescence plate reader. Magic Red has an optimal excitation at 592 nm and emission at 628 nm.
If working with adherent cells, please see the manual for additional protocols.
Product Specific References
| PMID | Publication |
| 38830624 | Lo, CH, et al. 2024. Acidic Nanoparticles Restore Lysosomal Acidification and Rescue Metabolic Dysfunction in Pancreatic β-Cells under Lipotoxic Conditions. ACS Nano, 15452-15467. |
| 38515365 | Sung, H.K., et al. 2024. Ischemia-induced cardiac dysfunction is exacerbated in adiponectin-knockout mice due to impaired autophagy flux. Clinical and translational science, e13758. |
| 37883971 | Ebner, M., et al. 2023. Nutrient-regulated control of lysosome function by signaling lipid conversion. Cell, . |
| 37142604 | Zeng, J., et al. 2023. Restoration of lysosomal acidification rescues autophagy and metabolic dysfunction in non-alcoholic fatty liver disease. Nature communications, 2573. |
| 37375535 | Zhang, C., et al. 2023. Bis-Benzylisoquinoline Alkaloids Inhibit Porcine Epidemic Diarrhea Virus by Disrupting Virus Entry. Pathogens (Basel, Switzerland). |
| 37417221 | Ezz, M.A., et al. 2023. Cathepsin L regulates oocyte meiosis and preimplantation embryo development. Cell proliferation, e13526. |
| 35535798 | Rodrigues, P.M., et al. 2022. LAMP2 regulates autophagy in the thymic epithelium and thymic stroma-dependent CD4 T cell development. Autophagy, 44940. |
| 35977928 | Ratto, E., et al. 2022. Direct control of lysosomal catabolic activity by mTORC1 through regulation of V-ATPase assembly. Nature communications, 4848. |
| 36535215 | Wu, K., et al. 2022. The endo-lysosomal regulatory protein BLOC1S1 modulates hepatic lysosomal content and lysosomal lipolysis. Biochemical and biophysical research communications, 44936. |
| 33372271 | Yang, Y., et al. 2021. Porphyromonas gingivalis facilitated the foam cell formation via lysosomal integral membrane protein 2 (LIMP2). Journal of periodontal research. |
| 33629936 | Wu, K., et al. 2021. BLOC1S1/GCN5L1/BORCS1 is a critical mediator for the initiation of autolysosomal tubulation. Autophagy. |
Question: When using a fluorescence plate reader staining for adherent cell, do I have to detach the cell by trypsin treatment? In that case, Is there anything to be aware of?
Answer: If you are using adherent cells, you do not necessarily need to detach them. If your plate reader is capable of reading from the bottom, you can grow your adherent culture in a tissue culture plate with black walls and a clear bottom.